Bayesian Survival Analysis with Data Augmentation
Motivation
When dealing with time-to-event data, right-censoring is a common occurance. Although most are familiar with likelihood construction under right-censoring (and corresponding frequentist estimation), there’s very little available online about Bayesian approaches even for fully parametric models. Here I’ll briefly outline a Bayesian estimation procedure for a Weibull model with right-censoring. The estimation procedure is MCMC based using a data augmentation approach.
As with most of my posts, all MCMC is coded from scratch. It helps me and it helps readers understand the underlying algorithm - an intuition that is more difficult to get if you’re just specifying the model in Stan.
Model Set Up
Suppose we observe
We’ll consider the setting where we regress on a binary treatment indicator,
From a Bayesian point of view, we are interested in the posterior
Data Augmentation
We’ll first look at the joint data distribution (the likelihood) for this problem. The central idea is to view the survival times for the
We also assume that subjects are independent so that
Now the integral is over the region
This is the usual likelihood for frequentist survival models: uncensored subjects contribute to the likelihood via the density while censored subjects contribute to the likelihood via the survival function R. For our Weibull model, it is 1-pweibull(). We would simply place priors on
But what if this integral was too hard to evaluate (as it may be for more complicated censoring mechanisms) and the complete data likelihood given below is easier?
Metropolis-in-Gibbs Sampler
The target posterior of interest is
The second conditional posterior is
The Gibbs sampler alternates between sampling from these two conditionals:
- Given parameters
- Combine these imputed values,
As the parameter estimates update, the imputations get better. As the imputations get better, the parameter estimates improve. Over time the process yields draws from the joint posterior
We retain the sample of
Simulation Example in R
All of the code implementing the augmented sampler (from scratch!) can be found on my GitHub. Basically I simulate a data set with a binary treatment indicator for 1,000 subjects with censoring and survival times independently drawn from a Weibull. \
For the
Here is the estimated survival function for each treatment group. Overlayed are the non-parametric estimates from a stratified Kaplan-Meier (KM) estimator. Note the parametric model is correctly specified here, so it does just as well as the KM in terms of estimating the mean curve. But the parametric model provides a less noisy fit - notice the credible bands are narrower at later time points when the at-risk counts get low in each treatment arm.
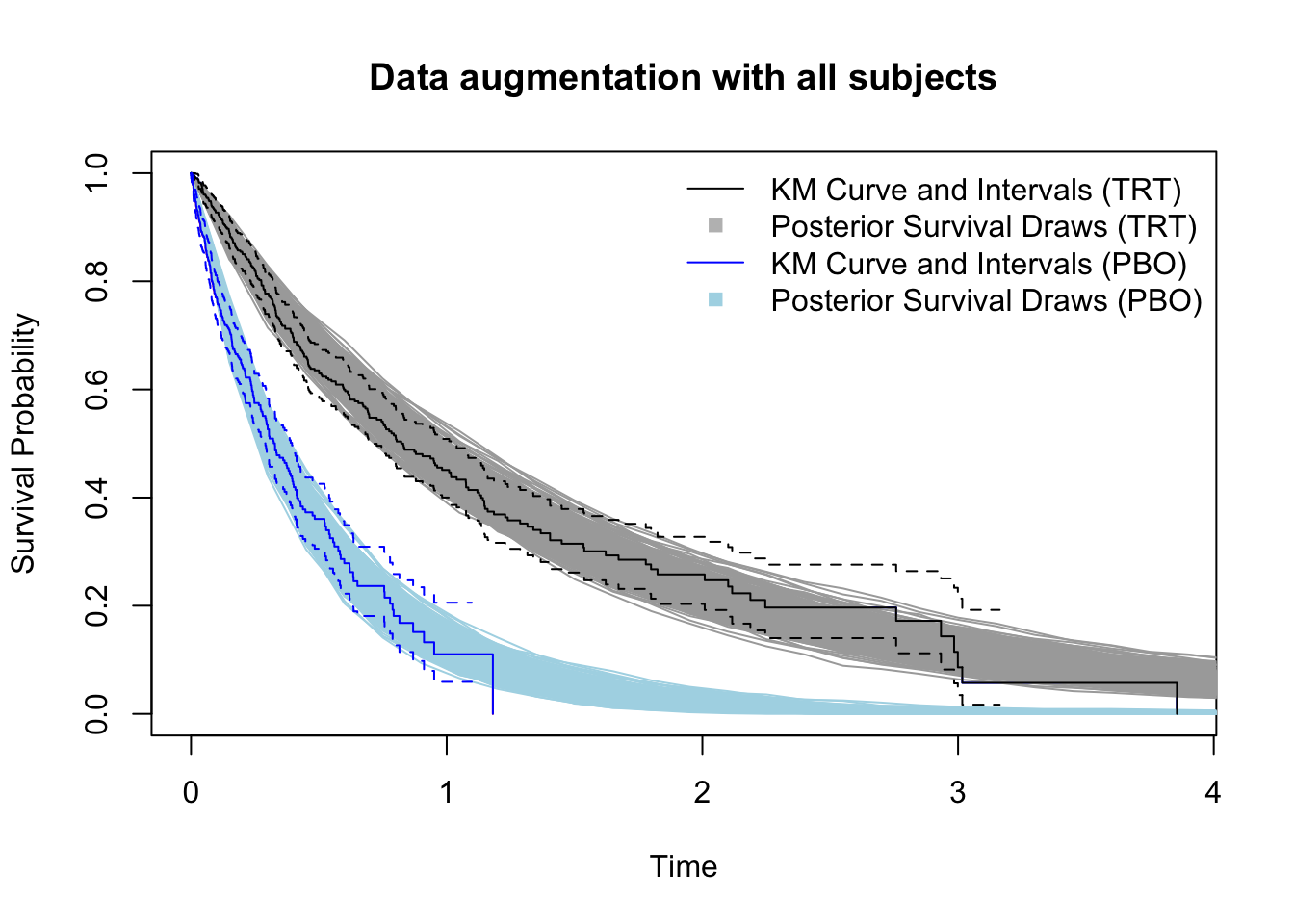
That’s just a helpful reminder of the efficiency gains parametric models have over nonparametric ones (when they’re correctly specified. Let’s take a look at the posterior distribution of the hazard ratio. The true value is indicated by the red line.
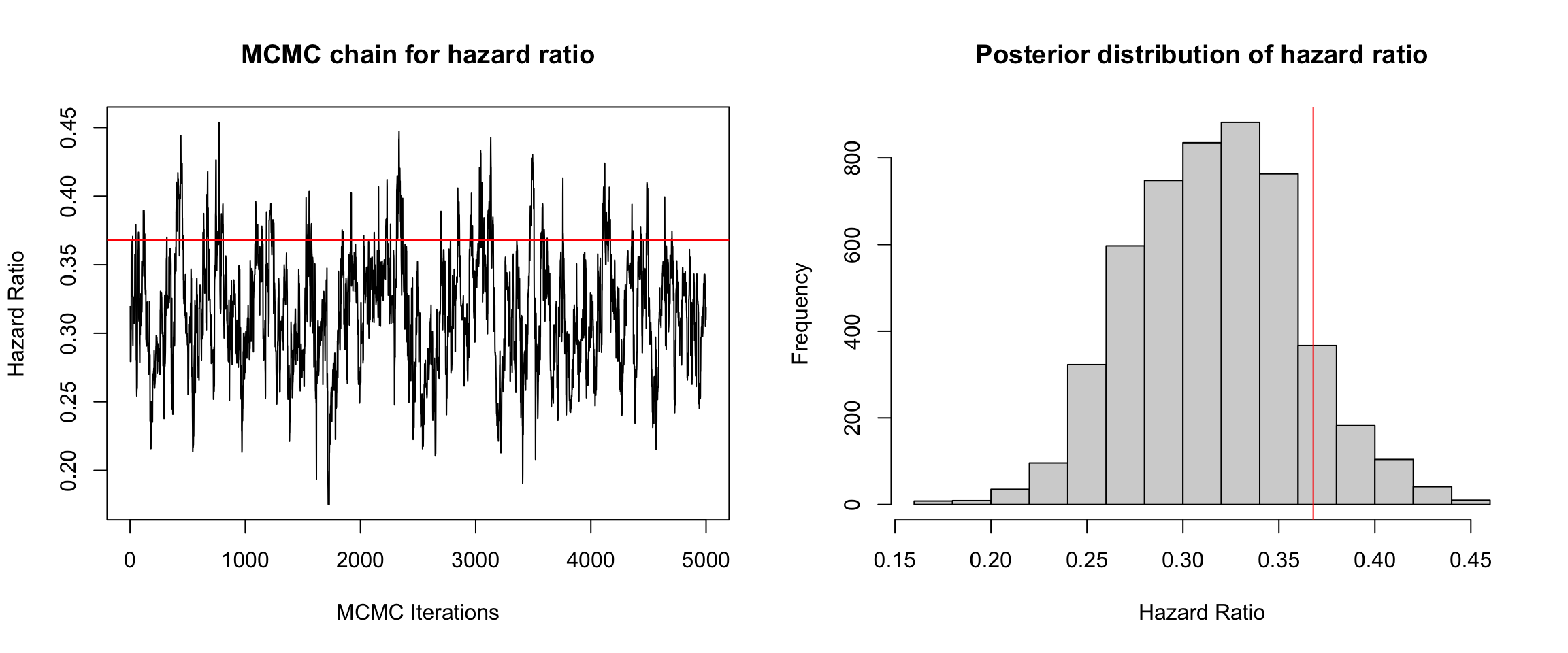 We could have run this thing for longer (and with multiple chains with different starting values). But I think this gets the point across. The posterior mean and
We could have run this thing for longer (and with multiple chains with different starting values). But I think this gets the point across. The posterior mean and